chip seq venn diagram
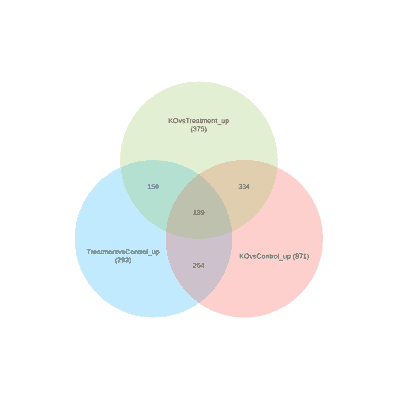
The Venn diagram on the right shows results from RNA-seq and ChiP-seq experiments designed to find Pax6 target genes as we discussed in class. We performed ChIP-seq analyses to explore genomic regions.
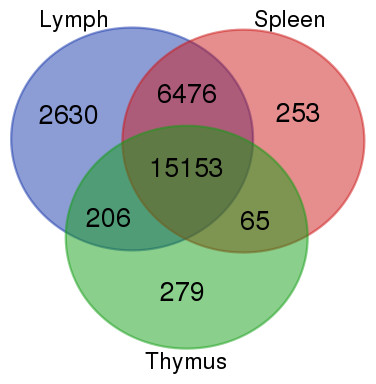
Rna Sequencing Rna Seq Of Lymph Node Spleen And Thymus Transcriptome From Wild Peninsular Malaysian Cynomolgus Macaque Macaca Fascicularis Peerj
Batch annotation of the peaks identified from either ChIP-seq ChIP-chip experiments or any experiments resulted in large number of chromosome ranges.

. Once peaks and TF binding sites are identified in ChIP-Seq data peaks can be annotated to the genome to find the nearest transcription start site. Compare the results with peaks identified. 51 Calculate overlap with genomic features like.
Workflow for ChIP-seq experiments of single transcription factor with replicates. A Venn diagram showing the overlap. 5 Functional annotation of ChIP-seq peaks.
The Venn diagram is not dawn to scale. ChIP-seq experiments are designed to isolate regions enriched in a factor of interest. Make Venn Diagram from two or more peak ranges Also calculate p-value to determine.
Workflow for ChIP-seq experiments of single transcription factor with replicates. The Venn diagram on the right shows results from RNA-seq and ChiP-seq. In total 559 genes 483 76 in Venn Diagram were found by RNA-seq to be differentially.
Going a step further I performed. B Read densities over ChIP-seq peaks of C11orf95fus1 and C11orf95-RELAfus1. A list of target genes generated from ChIP.
Contribute to Lie-neChIP-seq development by creating an account on GitHub. 44 Make a Venn-diagram showing the overlap of of ER and FOXA1 peaks. Examination of ChIP-seq and seq-DAP-seq bound regions and DE genes in the fourth whorl during carpel development reveals a high degree of overlap Figure 5 with not only many well.
The Venn diagram on the right shows results from RNA-seq and ChiP-seq experiments designed to find Pax6 target genes as we discussed in class. Venn Diagram of integrated ChIP-Seq and RNA-Seq results showing the direct and indirect targets of SMAD23 in the experiments. This workflow shows how to convert BEDGFF files to GRanges find overlapping peaks.
Call peaks with at least 2-3 software tools such as MACS2 slice coverage calling Bioc PeakSeq F-Seq Homer ChIPseqR or CSAR. The identification of enriched regions often refered to as peak finding is an area of research by. Transcription factors have two major.
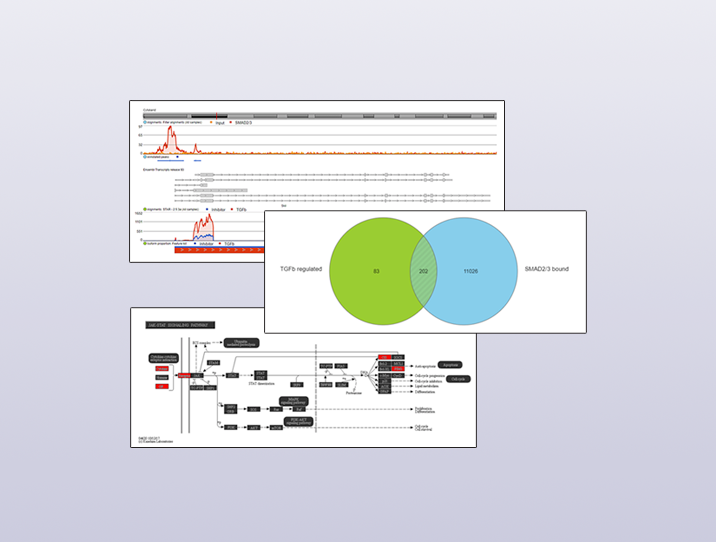
How To Integrate Chip Seq And Rna Seq Data Partek Inc
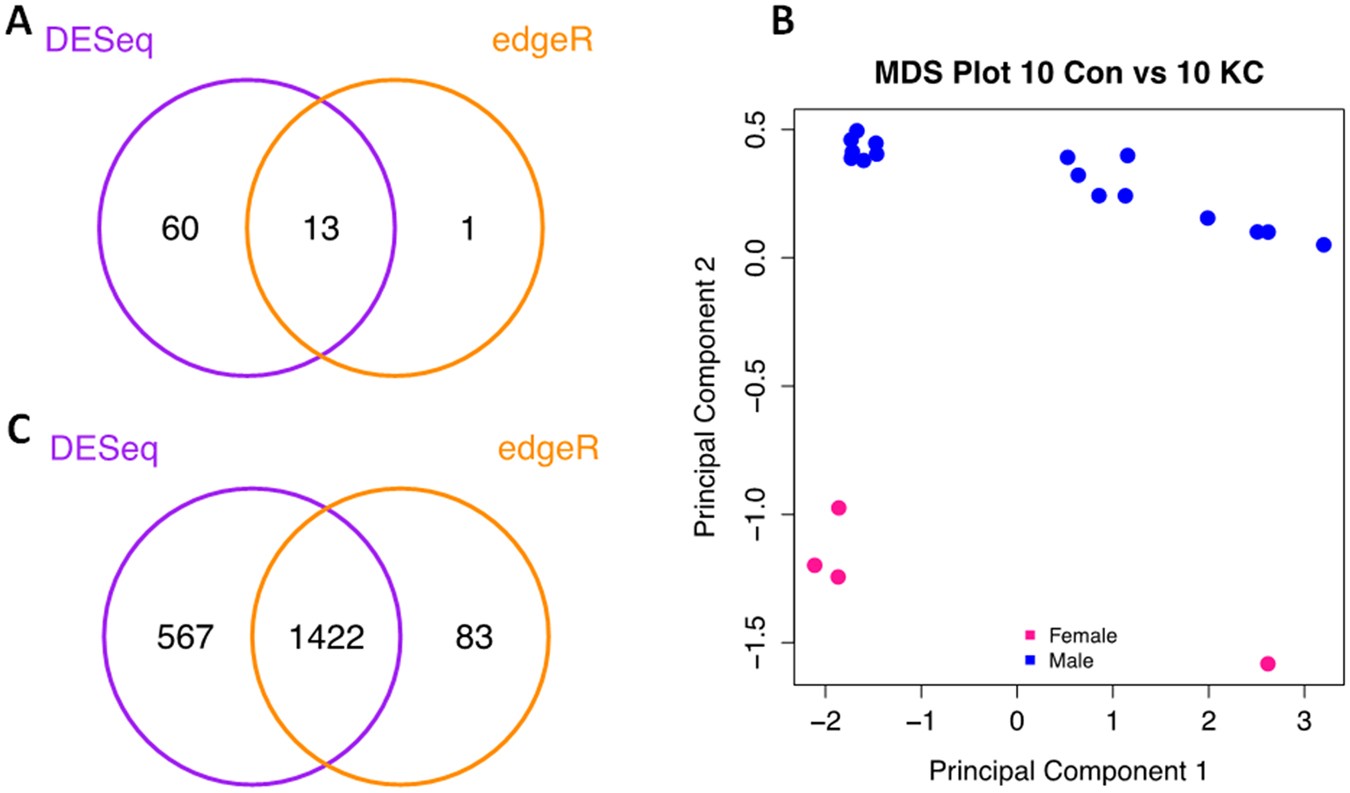
Rna Seq Analysis And Comparison Of Corneal Epithelium In Keratoconus And Myopia Patients Scientific Reports
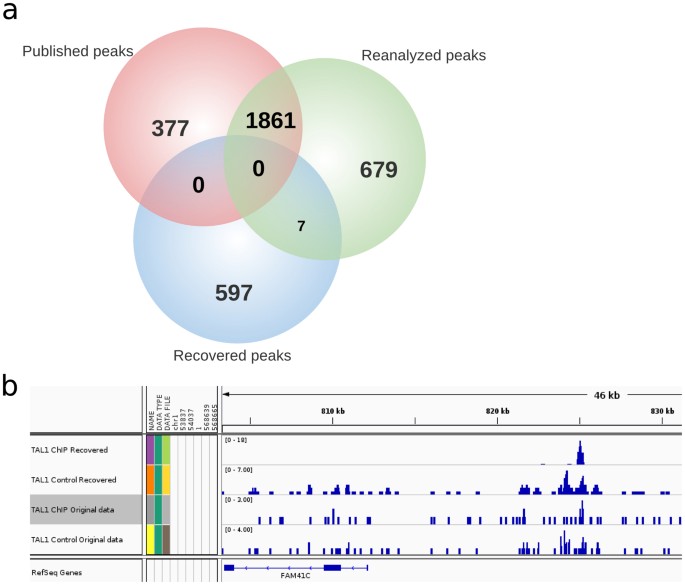
Important Biological Information Uncovered In Previously Unaligned Reads From Chromatin Immunoprecipitation Experiments Chip Seq Scientific Reports
A Widespread Distribution Of Genomic Cemyod Binding Sites Revealed And Cross Validated By Chip Chip And Chip Seq Techniques Plos One

A Venn Diagram Of The Chip Chip And Chip Seq Peaks Identified With Download Scientific Diagram
Gene Venn Diagram Tool Generate With Omicsbox

Phf2 Histone Demethylase Prevents Dna Damage And Genome Instability By Controlling Cell Cycle Progression Of Neural Progenitors Pnas

Coverage Plots Of H3k27ac Chip Seq Data In Breast Cancer Specimens Ppt Download
Figure 3 Unified Analysis Of Multiple Chip Seq Datasets Springerlink

A Venn Diagram Of Chip Seq Peaks The Dna Pull Down Were Done By Download Scientific Diagram

7 Rna Seq Data Ansysis Basic Scatter Plot Venn Diagram Etc Youtube

Afarp Chip Seq A Convenient And Reliable Method For Genome Profiling In As Few As 100 Cells With A Capability For Multiplexing Chip Seq Biorxiv

Venn Diagram Analysis Overestimates The Extent Of Circadian Rhythm Reprogramming Pelikan The Febs Journal Wiley Online Library

E Proteins Orchestrate Dynamic Transcriptional Cascades To Suppress Ilc2 Differentiation Biorxiv

Computational Methodology For Chip Seq Analysis
Loss Of The Caenorhabditis Elegans Pocket Protein Lin 35 Reveals Muvb S Innate Function As The Repressor Of Dream Target Genes Plos Genetics
Chromatin Enriched Rnas Mark Active And Repressive Cis Regulation An Analysis Of Nuclear Rna Seq Plos Computational Biology